Better predictions for better dairy cow breeding
Scientists from Agriculture Victoria, in collaboration with the University of Melbourne, have developed a world-first customised single nucleotide polymorphism (SNP) chip designed with the latest known genes that affect dairy cattle traits and therefore dairy cow breeding.
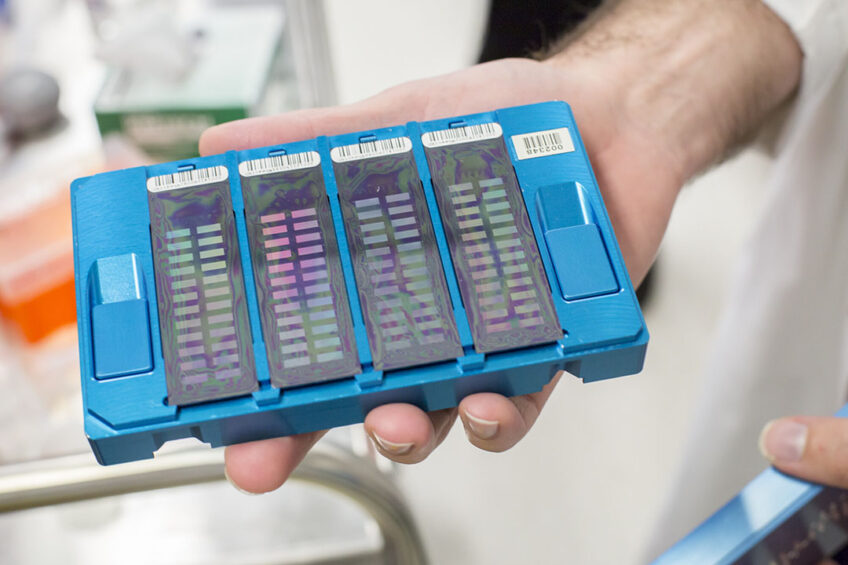
The work involves the customisation of a small piece of plastic – an SNP chip – with 50,000 small dots that bind DNA to detect genetic variations on the bovine genome, with the potential to improve dairy breeding worldwide.
The information obtained from an SNP chip can be used to make genetic predictions for a trait of interest, so breeders can quickly narrow down which animals they are interested in breeding.
Agriculture Victoria research scientist and research fellow with the University of Melbourne, Dr Ruidong Xiang, says SNP chips help breeders select animals with genetics of interest by testing tens of thousands of genetic variants to find the genes for important traits in each animal for the purposes of breeding.
“The information obtained from an SNP chip can be used to make genetic predictions for a trait of interest, such as milk production, so breeders can quickly narrow down which animals they are interested in breeding,” he explains.
Better predictions to deliver good improvements
The research team started with 16 million genomic variants from 44,000 Australian dairy cattle and moved along the bovine genome, picking the next best variant until they narrowed it down to 50,000 genetic variations for inclusion on the SNP chip.
The new customised SNP chip will have better predictions for a trait of interest compared to existing chips currently used in the industry and will ultimately improve breeding decisions. “Existing chips are not optimal,” Dr Xiang says. “But these chips are routinely used to test cows, so it is an important basic tool. Improving the chips delivers some good improvements to the dairy industry.”
Dr Xiang says that there was room to improve existing chips by using better markers. “So, we carried on and did that.” The new method includes using more genes associated with 37 dairy traits. This is a huge improvement on the standard chips, as most genetic variants used in the standard chip were not supported by biology.
“The genetic markers on the standard chip are not supported by the biology of traits,” Dr Xiang says. “They are still selected based on factors like the presence of markers in different breeds and the density of markers in the cow genome. However, they are not selected based on the biology, such as if they affect dairy traits, change different cells, tissues or metabolism.”
Long-term benefit for dairy cow breeding
The aim of the new chip is to include genetic markers that affect dairy traits and that play a role in regulating cellular processes or metabolism. “We have done a lot of work to find those important markers and then we put them on the new chip, and that is why it is biology-informed,” Dr Xiang points out.
The research findings showed the customised biology-informed chip outperformed the standard chip in predicting the genetic value of multiple traits across datasets from 90,000 dairy cattle by up to 10%.
Dr Xiang: “Outperforming the standard chip by 10% doesn’t sound like a lot, but it is a permanent and accumulated gain for improving the genetics of Australian major dairy breeds, such as Holstein, Jersey and Aussie Red. Once the genetics are improved, it won’t go away. The benefit of having cows with better genetics will be permanent and will keep improving from generation to generation of more accurate selection. Therefore, the benefit for Australian dairy farmers will be consistent and long term.”

The research team eventually used the datasets of 70,000 Australian dairy cattle and about 20,000 cattle from the United States. “So that gives improvements for the cattle in several countries,” Dr Xiang says.
Agriculture Victoria Research’s high throughput sequencing capabilities and the Biosciences Advanced Scientific Computing facility at AgriBio were used to analyse the vast amount of data. They were also used to develop the methodology and statistical methods utilised in the research, such as incorporating Bayesian modelling.
15,000 cows tested
Dr Xiang says that by translating big genomic data, the research team has been able to develop an industry-usable product that will help farmers select and breed the best animals more accurately and efficiently.
Agriculture Victoria has a pilot project allowing some farmers to have access to the new chip, which benefits farmers from the top down. Most Australian dairy cows will be genetically evaluated by national facilities (like DataGene), and the results will be fed back to farmers. The new SNP chip will be used in a national genetic evaluation of cows; therefore, soon all Australian dairy farmers will benefit from the new chip.
Using the new chip, the national facility has started evaluating Australian Red cattle. Farmers working with Australian Red are the first to benefit; this will be expanded to Holstein and Jersey cows in the foreseeable future.
“Quite a few cows have already been genotyped with the new chip,” Dr Xiang says. “Around 15,000 dairy cows in Australia have already been tested. There are many Australian cows, but for now mainly Australian Red cattle have been tested. And small numbers of Holstein and Jersey cows. They are the next target to apply the new chip on. We are doing further analysis on our results.” Dr Xiang expects all dairy breeds in Australia can eventually be tested with the new chip.

Dr Xiang emphasises that the research has the potential to benefit the Australian dairy industry as well as other key global dairy regions. The new chip was tested in predicting performances of dairy cattle in the US and New Zealand. There, test results are also indicating that the new chip predicted performances more accurately than the standard chip.
“We have a collaboration with scientists in other countries,” Dr Xiang says. “We share the scientific information on these chips. Each marker is important. Other countries will develop their own chip. Through collaboration they can have an understanding of what is working in our chip and improve on our work. Some New Zealand companies, for example, use our information to improve on their design.”
Testing cows, and animal ranking
For cows to be tested, farmers have to send tail hair of their animals to the national testing centre. “That minimises the damage to the animal,” Dr Xiang says. “In Victoria, farmers would send it to DataGene. The first step would be to measure the genes and compare the data to all the animals that are in the database. We can then give an animal ranking based on genetic merit. Based on the information we have, we can give a prediction of how good a cow is.”
With the improved chip, the prediction of the animal will get better. “But it’s like a weather forecast,” Dr Xiang says. “It is not 100% accurate.”
The research of Dr Xiang is part of the DairyBio Animal Program, which is targeting an additional value of AUS$ 350 per cow per year in Australian dairy herds through genetic improvement, lower costs by enabling selection for health traits and developing improved breeding management tools.
The research aligns with the Grow, Modernise and Promote themes of the Victorian Agriculture Strategy. DairyBio is a research and innovation initiative of Dairy Australia, Gardiner Foundation and Agriculture Victoria Research, further supported by the Australian Research Council and in collaboration with Dairy New Zealand.
More about the research findings can be found in the open access scientific journal, Nature Communications.
Join 13,000+ subscribers
Subscribe to our newsletter to stay updated about all the need-to-know content in the dairy sector, two times a week.